Overview
This gfcanalysis R package facilitates simple analyses using the Hansen et
al. 20131 Global Forest Change
dataset.
The package was written to analyze forest change in within the Zone of
Interaction surrounding each of the forest monitoring sites of the Tropical
Ecology Assessment and Monitoring (TEAM) Network.
If you need help with any of the functions in the package, see the help files
for more information. For example, type ?download_tiles in R to see the help
file for the download_tiles function.
Getting started
This post will outline an analysis using the gfcanalysis package. Note that
as the computations are intensive, some parts of this analysis may take some
time to run (about 30 minutes total to run all of the code outlined here). If
you do not already have the GFC product data downloaded on your computer,
downloading the dataset will also take some time (though this process is
automated by gfcanalysis).
To get started, first install the gfcanalysis package from CRAN. Also install
the rgdal package needed for reading/writing shapefiles if you do not already
have it.
if (!require(gfcanalysis)) install.packages('gfcanalysis')## Loading required package: gfcanalysis
## Loading required package: raster
## Loading required package: spif (!require(rgdal)) install.packages('rgdal')## Loading required package: rgdal
## rgdal: version: 0.8-16, (SVN revision 498)
## Geospatial Data Abstraction Library extensions to R successfully loaded
## Loaded GDAL runtime: GDAL 1.10.1, released 2013/08/26
## Path to GDAL shared files: C:/Users/azvoleff/R/win-library/3.2/rgdal/gdal
## GDAL does not use iconv for recoding strings.
## Loaded PROJ.4 runtime: Rel. 4.8.0, 6 March 2012, [PJ_VERSION: 480]
## Path to PROJ.4 shared files: C:/Users/azvoleff/R/win-library/3.2/rgdal/projIndicate where we want to save GFC tiles downloaded from Google. For any given AOI, the script will first check to see if these tiles are available locally (in the below folder) before downloading them from the server - so I recommend storing ALL of your GFC tiles in the same folder. For this example we will use “.” - the current working directory of the R session.
output_folder <- "."Set the threshold for forest/non-forest based on the treecover2000 layer in the GFC product:
forest_threshold <- 90Downloading data from Google server for a given AOI
Load an area of interest. For this example we use a shapefile of the Zone of Interaction (ZOI) of the TEAM Network site in Nam Kading National Protected Area, Laos. Notice that first we specify the folder the shapefile is in (here it is a “.” indicating the current working directory), and then the name of the shapefile without the “.shp”. To follow along with this example, download this shapefile of the ZOI.
aoi <- readOGR('.', 'ZOI_NAK_2012_EEsimple')## OGR data source with driver: ESRI Shapefile
## Source: ".", layer: "ZOI_NAK_2012_EEsimple"
## with 1 features and 3 fields
## Feature type: wkbPolygon with 2 dimensionsCalculate the tiles needed to cover the AOI:
tiles <- calc_gfc_tiles(aoi)
print(length(tiles)) # Number of tiles needed to cover AOI## [1] 1To check the overlap between the tiles and the aoi, you can make a plot of the needed tiles and the AOI using R’s plotting functions:
plot(tiles)
plot(aoi, add=TRUE, lty=2, col="#00ff0050")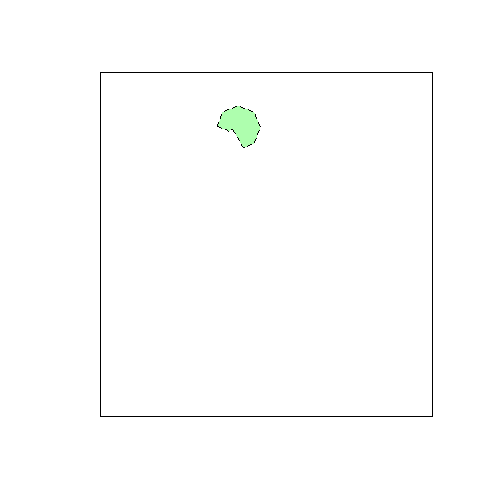
Now, check to see if these tiles are already present locally, and download them
if they are not. By default the “first” and “last” composite surface
reflectance images are not downloaded. To also download these images specify
first_and_last=TRUE.
download_tiles(tiles, output_folder, first_and_last=FALSE)## 1 tiles to download/check.
## 0 file(s) succeeded, 5 file(s) skipped, 0 file(s) failed.Performing thresholding and calculating basic statistics
Extract the GFC data for this AOI from the downloaded GFC tiles, mosaicing multiple tiles as necessary (if needed to cover the AOI). Save this extract in GeoTiff format in the current working directory (can also save as ENVI format, Erdas format, etc.)
gfc_extract <- extract_gfc(aoi, output_folder, filename="NAK_GFC_extract.tif")The extracted dataset has 5 layers (not yet thresholded):
gfc_extract## class : RasterBrick
## dimensions : 4358, 4761, 20748438, 5 (nrow, ncol, ncell, nlayers)
## resolution : 0.0002778, 0.0002778 (x, y)
## extent : 103.5, 104.8, 17.83, 19.04 (xmin, xmax, ymin, ymax)
## coord. ref. : +proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0
## data source : C:\Users\azvoleff\Code\Misc\azvoleff.github.io\Rmd\2014-03-25-analyzing-forest-change-with-gfcanalysis\NAK_GFC_extract.tif
## names : treecover2000, loss, gain, lossyear, datamask
## min values : 0, 0, 0, 0, 1
## max values : 100, 1, 1, 12, 2Threshold the GFC data based on a specified percent cover threshold (0-100), and save the thresholded layers to a GeoTiff:
gfc_thresholded <- threshold_gfc(gfc_extract, forest_threshold=forest_threshold,
filename="NAK_GFC_extract_thresholded.tif")Coding of the thresholded output
The thresholded dataset has 5 layers:
gfc_thresholded## class : RasterBrick
## dimensions : 4358, 4761, 20748438, 5 (nrow, ncol, ncell, nlayers)
## resolution : 0.0002778, 0.0002778 (x, y)
## extent : 103.5, 104.8, 17.83, 19.04 (xmin, xmax, ymin, ymax)
## coord. ref. : +proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0
## data source : C:\Users\azvoleff\Code\Misc\azvoleff.github.io\Rmd\2014-03-25-analyzing-forest-change-with-gfcanalysis\NAK_GFC_extract_thresholded.tif
## names : forest2000, lossyear, gain, lossgain, datamask
## min values : 0, 0, 0, 0, 1
## max values : 1, 12, 1, 1, 2The output is coded using the following coding scheme:
Band 1 (forest2000)
Based on the provided forest_threshold. Pixels wiwth percent canopy cover
greater than forest_threshold are coded as forest.
| Cover in 2000 | Code |
|---|---|
| Non-forest | 0 |
| Forest | 1 |
Band 2 (lossyear)
Note that lossyear is zero for pixels that were not forested in 2000.
| Year of loss | Code |
|---|---|
| No loss | 0 |
| Loss in 2001 | 1 |
| Loss in 2002 | 2 |
| Loss in 2003 | 3 |
| Loss in 2004 | 4 |
| Loss in 2005 | 5 |
| Loss in 2006 | 6 |
| Loss in 2007 | 7 |
| Loss in 2008 | 8 |
| Loss in 2009 | 9 |
| Loss in 2010 | 10 |
| Loss in 2011 | 11 |
| Loss in 2012 | 12 |
Band 3 (gain)
Note that gain is zero for pixels that were forested in 2000.
| Change | Code |
|---|---|
| No gain | 0 |
| Gain | 1 |
Band 4 (lossgain)
Note that loss and gain is difficult to interpret from the thresholded
product, as the original GFC product does not provide information on the
sequence of loss and gain (loss then gain, or gain then loss). The product also
does not provide information on the levels of canopy cover reached prior to
loss (for gain then loss) or after loss (for loss then gain pixels). The layer
is calculated here as: lossgain <- gain & (lossyear != 0), where lossyear
and gain are the original GFC gain and lossyear layers, respectively.
| Change | Code |
|---|---|
| No loss and gain | 0 |
| Loss and gain | 1 |
Band 5 (datamask)
| Class | Code |
|---|---|
| No data | 0 |
| Land | 1 |
| Water | 2 |
Calculating statistics on forest loss and forest gain
Calculate annual statistics on forest loss/gain:
gfc_stats <- gfc_stats(aoi, gfc_thresholded)## Data appears to be in latitude/longitude. Calculating cell areas on a sphere.gfc_stats## $loss_table
## year aoi cover loss
## 1 2000 AOI 1 433656 NA
## 2 2001 AOI 1 433125 530.6
## 3 2002 AOI 1 432108 1017.4
## 4 2003 AOI 1 430451 1656.4
## 5 2004 AOI 1 429590 861.6
## 6 2005 AOI 1 427738 1851.3
## 7 2006 AOI 1 425938 1800.2
## 8 2007 AOI 1 421196 4742.1
## 9 2008 AOI 1 420032 1164.0
## 10 2009 AOI 1 415982 4049.5
## 11 2010 AOI 1 412196 3786.5
## 12 2011 AOI 1 407462 4734.3
## 13 2012 AOI 1 403578 3884.1
##
## $gain_table
## period aoi gain lossgain
## 1 2000-2012 AOI 1 16194 12287Save these statistics to CSV files for use in Excel, or other software:
write.csv(gfc_stats$loss_table,
file='NAK_GFC_extract_thresholded_losstable.csv', row.names=FALSE)
write.csv(gfc_stats$gain_table,
file='NAK_GFC_extract_thresholded_gaintable.csv', row.names=FALSE)To view the output format of the CSV files output by gfcanalysis, see the
loss
table
and gain
table
for Nam Kading.
Making simple visualizations
There is also a function in gfcanalysis to calculate and save a thresholded
annual layer stack from the GFC product (useful for simple visualizations,
etc.):
gfc_annual_stack <- annual_stack(gfc_thresholded)
writeRaster(gfc_annual_stack, filename="NAK_GFC_extract_thresholded_annual.tif")## class : RasterBrick
## dimensions : 4358, 4761, 20748438, 13 (nrow, ncol, ncell, nlayers)
## resolution : 0.0002778, 0.0002778 (x, y)
## extent : 103.5, 104.8, 17.83, 19.04 (xmin, xmax, ymin, ymax)
## coord. ref. : +proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0
## data source : C:\Users\azvoleff\Code\Misc\azvoleff.github.io\Rmd\2014-03-25-analyzing-forest-change-with-gfcanalysis\NAK_GFC_extract_thresholded_annual.tif
## names : NAK_GFC_e//d_annual.1, NAK_GFC_e//d_annual.2, NAK_GFC_e//d_annual.3, NAK_GFC_e//d_annual.4, NAK_GFC_e//d_annual.5, NAK_GFC_e//d_annual.6, NAK_GFC_e//d_annual.7, NAK_GFC_e//d_annual.8, NAK_GFC_e//d_annual.9, NAK_GFC_e//_annual.10, NAK_GFC_e//_annual.11, NAK_GFC_e//_annual.12, NAK_GFC_e//_annual.13
## min values : 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1
## max values : 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6The annual stack output by annual_stack has one layer for each year:
gfc_annual_stack## class : RasterBrick
## dimensions : 4358, 4761, 20748438, 13 (nrow, ncol, ncell, nlayers)
## resolution : 0.0002778, 0.0002778 (x, y)
## extent : 103.5, 104.8, 17.83, 19.04 (xmin, xmax, ymin, ymax)
## coord. ref. : +proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0
## data source : C:\Users\azvoleff\AppData\Local\Temp\R_raster_azvoleff\raster_tmp_2014-04-04_141034_13628_76953.grd
## names : y2000, y2001, y2002, y2003, y2004, y2005, y2006, y2007, y2008, y2009, y2010, y2011, y2012
## min values : 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1
## max values : 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6
## time : 2000-01-01, 2001-01-01, 2002-01-01, 2003-01-01, 2004-01-01, 2005-01-01, 2006-01-01, 2007-01-01, 2008-01-01, 2009-01-01, 2010-01-01, 2011-01-01, 2012-01-01Forest change in each year is coded as:
| Cover/Change | Code |
|---|---|
| No data | 0 |
| Forest | 1 |
| Non-forest | 2 |
| Forest loss | 3 |
| Forest gain | 4 |
| Forest loss and gain | 5 |
| Water | 6 |
The animate_annual function can be used to save a simple visualization of the
thresholded annual layer stack.
Note: For this example, we are using the data in the WGS84 coordinate system.
For a real analysis or presentation, the data should be projected into UTM or
another projection system for this. The utm_zone function in the
gfcanalysis package and the projectRaster function in the raster package
could be used to automate this. Also see the to_utm option for the
extract_gfc function (type ?extract_gfc in R).
To make an annual animation (in WGS84) type:
aoi$label <- "ZOI" # Label the polygon on the plot
animate_annual(aoi, gfc_annual_stack, out_dir='.', site_name='Nam Kading')## HTML file created at: C:\Users\azvoleff\Code\Misc\azvoleff.github.io\Rmd\2014-03-25-analyzing-forest-change-with-gfcanalysis/gfc_animation.html
## You may use ani.options(outdir = getwd()) or saveHTML(..., outdir = getwd()) to generate files under the current working directory.The animation will be saved in the directory specified by out_dir (in this
example the current working directory). To view the animation, double-click the
new “.html” file in that directory. The animation will look something like
this.
-
Hansen, M. C., P. V. Potapov, R. Moore, M. Hancher, S. A. Turubanova, A. Tyukavina, D. Thau, S. V. Stehman, S. J. Goetz, T. R. Loveland, A. Kommareddy, A. Egorov, L. Chini, C. O. Justice, and J. R. G. Townshend. 2013. High-Resolution Global Maps of 21st-Century Forest Cover Change. Science 342, (15 November): 850–853. Data available on-line from: http://earthenginepartners.appspot.com/science-2013-global-forest. ↩
